Molecule.
I am fascinated by the molecular mechanisms that drive plant development. The interplay of small molecules—such as DNA, RNA, proteins, and metabolites—can lead to the emergence of complex plant morphology. Currently, I mainly use single-cell and spatial transcriptomics to unravel the molecular processes governing rice root development and its responses to soil stresses.
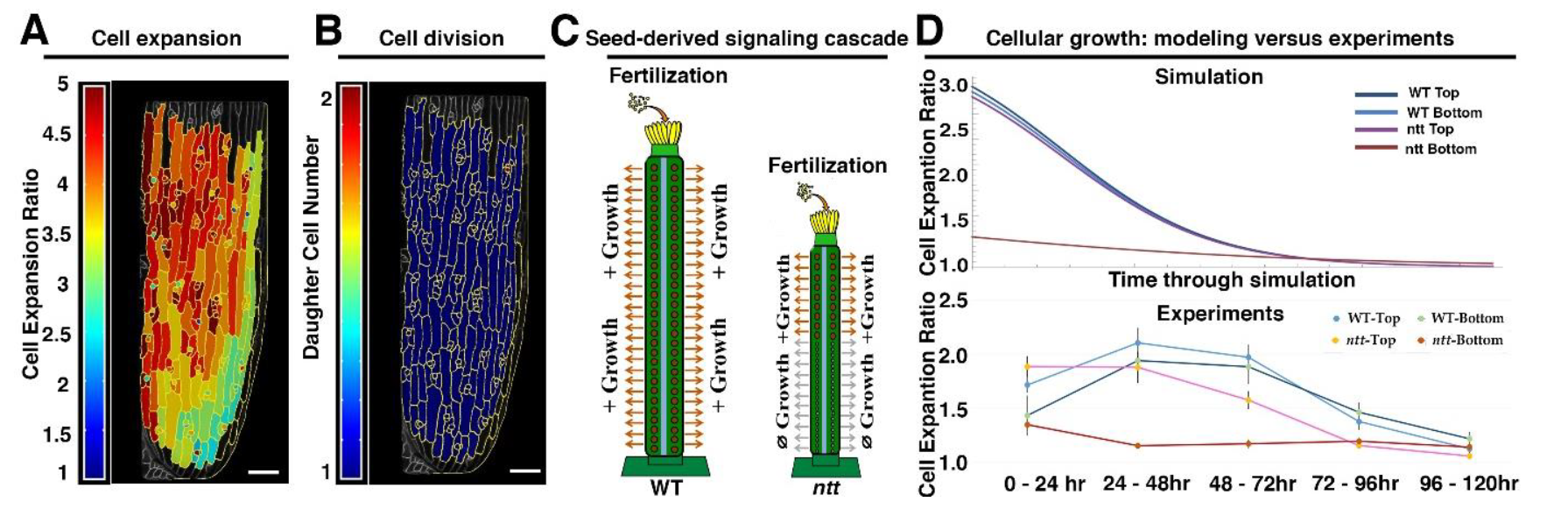
During my PhD in Dr. Adrienne Roeder’s lab, I explored the connection between individual cell behaviors and overall organ growth, with a focus on post-fertilization fruit development in Arabidopsis. I developed a high-resolution live-imaging platform specifically designed to study fruit growth dynamics. Collaborating with Dr. Arezki Boudaoud, I discovered the transformative power of combining computational modeling with quantitative growth analysis. This integrative approach revealed how cell growth is finely tuned by a seed-derived signaling cascade (Ripoll*, Zhu* et al., 2019 PNAS).
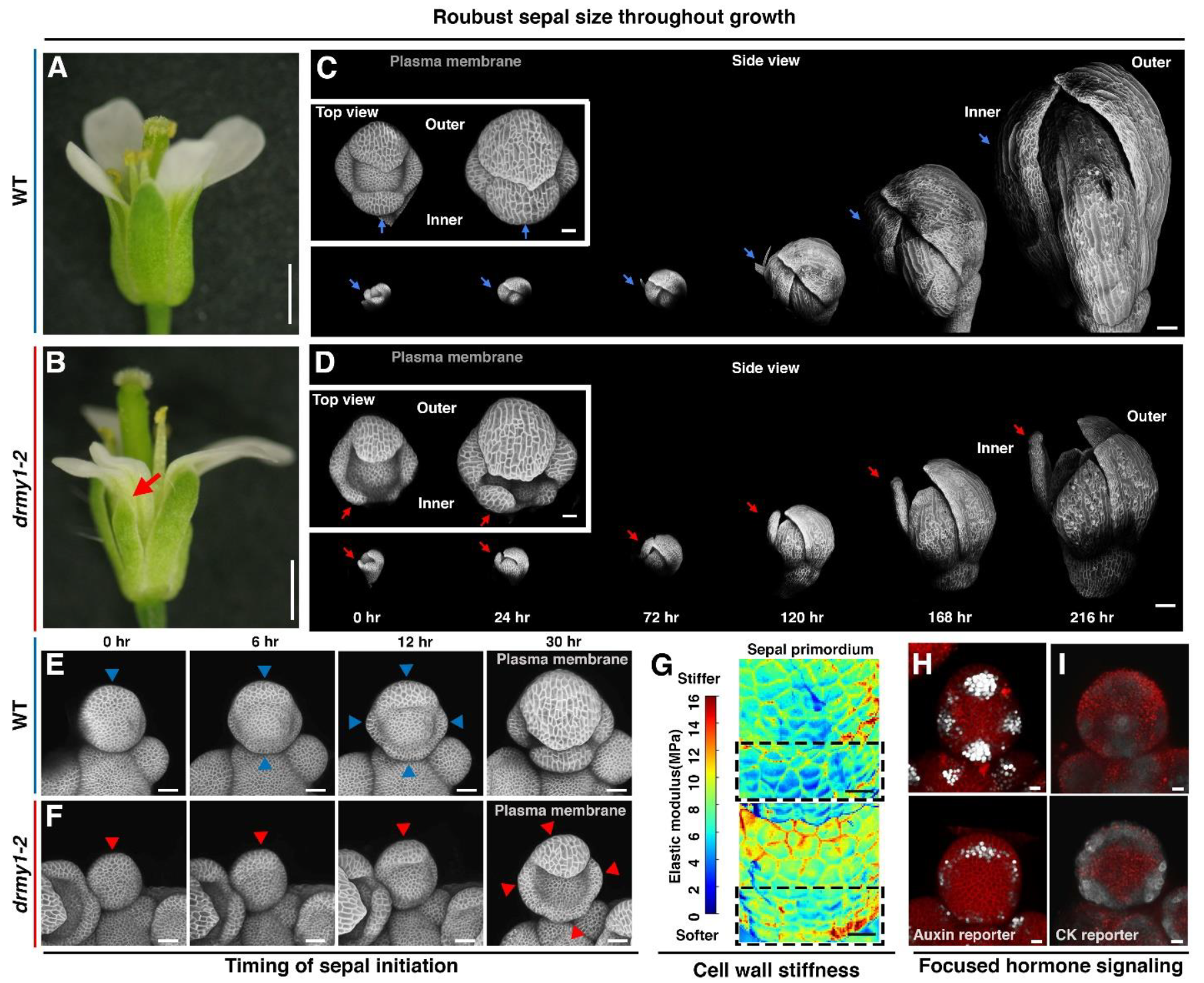
I also investigated the robustness of Arabidopsis sepal growth, which is essential for protecting inner floral organs. Through extensive use of live imaging, combined with transcriptomic and proteomic analyses, I identified coordinated patterns of auxin and cytokinin accumulation, as well as the precise regulation of protein homeostasis, as the key factors controlling sepal development. These factors collectively orchestrate cellular growth and establish robust sepal initiation patterns (Zhu et al., 2020 Nature Plants; Kong*, Zhu* et al., 2024 Developmental Cell).
I utilized scRNA-seq and spatial transcriptomics to establish a comprehensive single-cell RNA sequencing reference atlas for rice primary roots, providing a solid foundation for future rice scRNA-seq studies. I also pioneered the application of scRNA-seq and spatial transcriptomics to rice roots grown in soils. I found that soil compaction upregulated the expression of genes triggering water impermeable barrier formation in outer root (exodermis) tissues. Such upregulation was induced by enhanced ABA biosynthesis and release from inner (phloem) tissues. Our single cell resolution study revealed how root tissues coordinate and adapt to soil compaction conditions (Zhu*, Hsu* et al., 2024 Under revision)
