Molecule.
I am fascinated by the molecular mechanisms that drive plant development. The interplay of small molecules—such as DNA, RNA, proteins, and metabolites—can lead to the emergence of complex plant morphology.
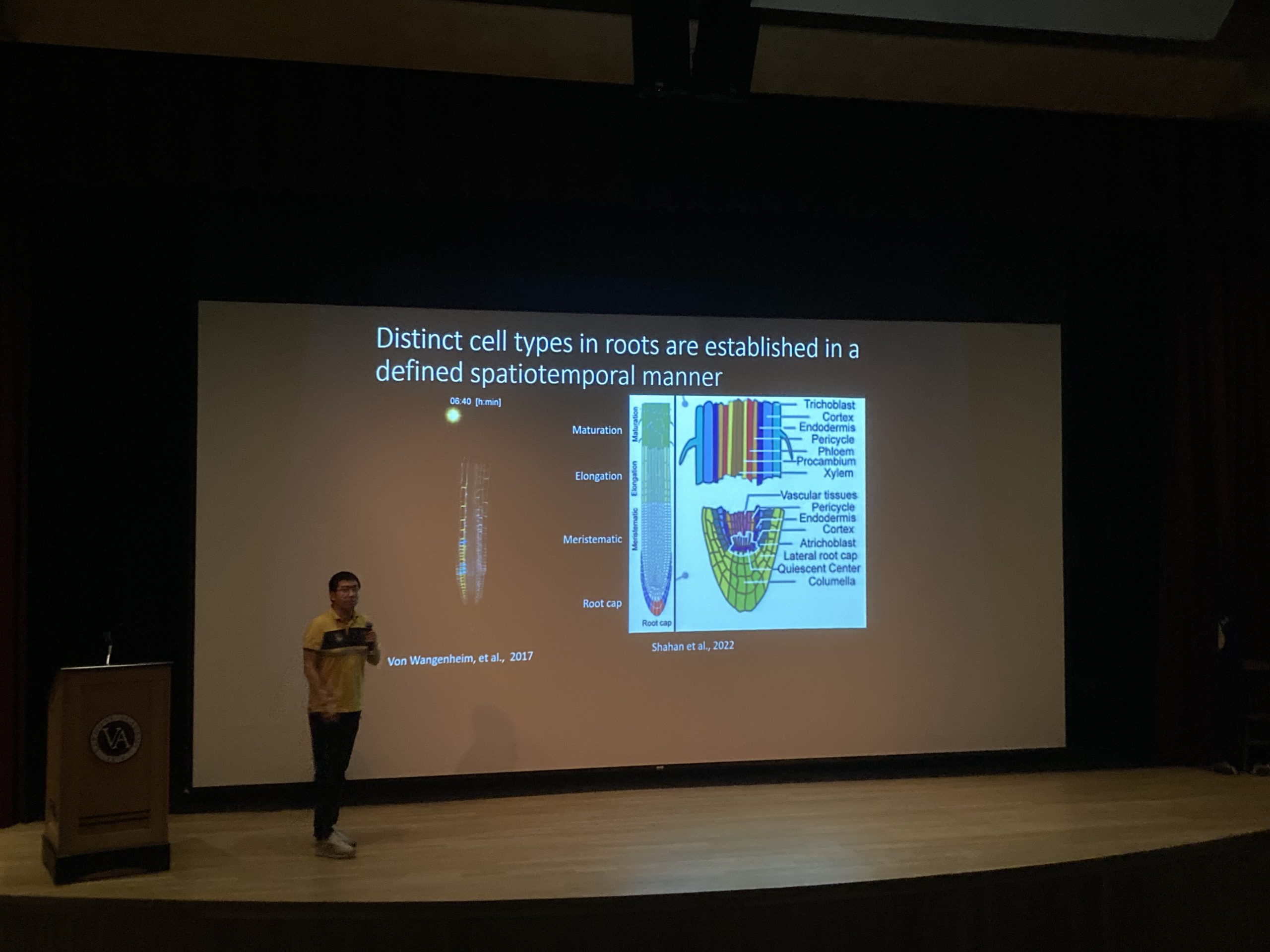
Currently, I mainly use single-cell and spatial transcriptomics to unravel the molecular processes governing rice root development and its responses to soil stresses.